Research Projects
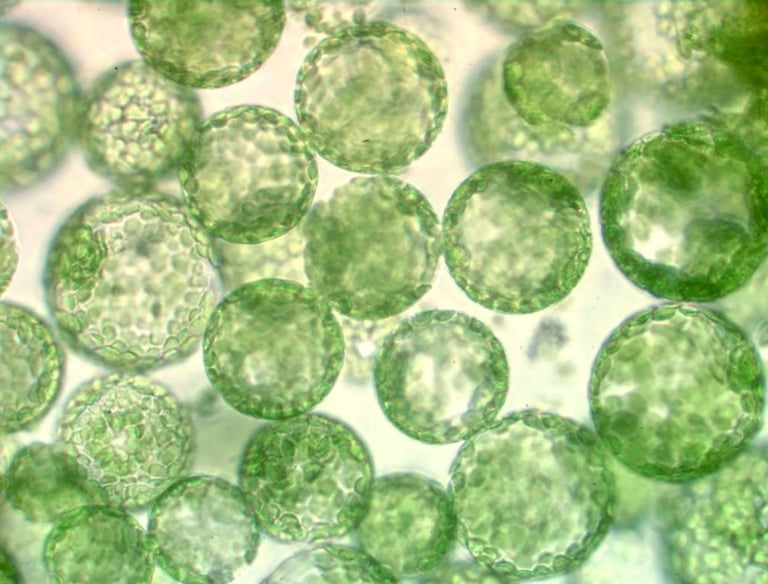
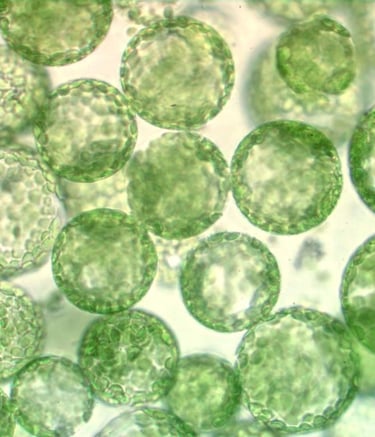
Current Interest - Dedifferentiation of Plant Cells In vitro
Despite the wide use of cell/tissue culture and regeneration procedures, there are numerous species, genotypes, and explant types that are notoriously recalcitrant for dedifferentiation and cell cycle reactivation. These include the most important cereal species, where only young tissues of certain genotypes are amenable to in vitro culture and regeneration. Most often, embryogenic cell cultures of specific genotypes have to be established and maintained to have a sufficient amount of cells for biotechnological applications. The direct use of their differentiated mesophyll cells for this purpose, which is possible in many dicot species, would be very useful, especially in the era of genome editing. Among others, transfection of cereal leaf cells/protoplasts with constructs expressing Cas9 nuclease and guide RNAs allows targeted genome editing without transgene insertion. However, the bottleneck is the subsequent plant regeneration that is strongly genotype-dependent and prevents the direct application of the technology for most of the high-yielding cultivars. Parallel transient expression of genes overriding the barriers in dedifferentiation or cell cycle progression could solve this problem.
Understanding what causes the recalcitrance of cereal somatic plant cells to in vitro culture conditions (especially wounding and hormones) could allow us to widen the available germplasm for plant biotechnology applications, besides gaining insights into the flexible cell differentiation strategy of plants. Our research pioneers the investigation of the role of the cell division/differentiation regulator DREAM protein complexes during in vitro plant regeneration (both in a dicot and a monocot), as well as the identification of cereal DREAM complexes. The combination of the set of our previously collected/produced Arabidopsis mutants/transgenic lines of DREAM complex components and our long-standing experience in in vitro plant cell culture and regeneration provides us an unprecedented possibility to answer the long-standing yet unanswered scientific question: how a differentiated somatic plant cell can regain its developmental potential.
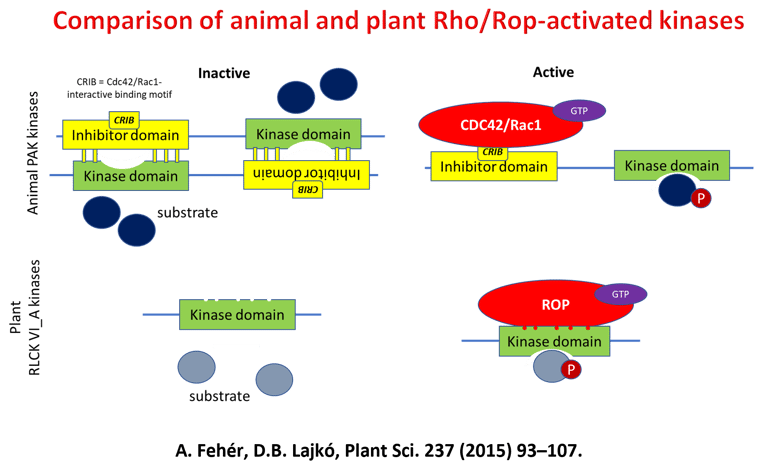
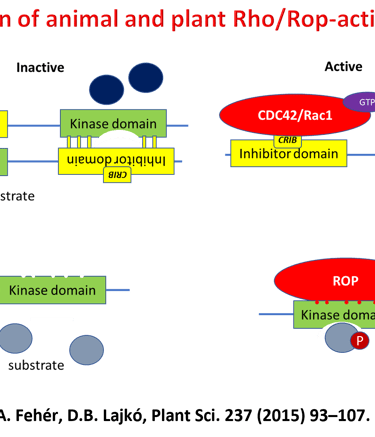


Earlier Project - Kinase Signalling Downstream of ROP GTPases
The structure of the plant body is fundamentally determined by the direction of cell division, shape, and growth during plant development. The direction of cell division and growth is regulated by similar molecular processes in most eukaryotic organisms. At the heart of this regulation are Rho-type GTP-binding proteins. These proteins function as two-position molecular switches due to their GTP-binding and hydrolyzing activities (GTP binding = in, GTP hydrolysis = out). In cells, the ratio of GTP- and GDP-bound (i.e. switched on or off) forms of Rho GTPase is regulated by proteins specialized for this task. When turned on, GTPase can interact specifically with effector proteins that, when activated, trigger a response to the given signal.
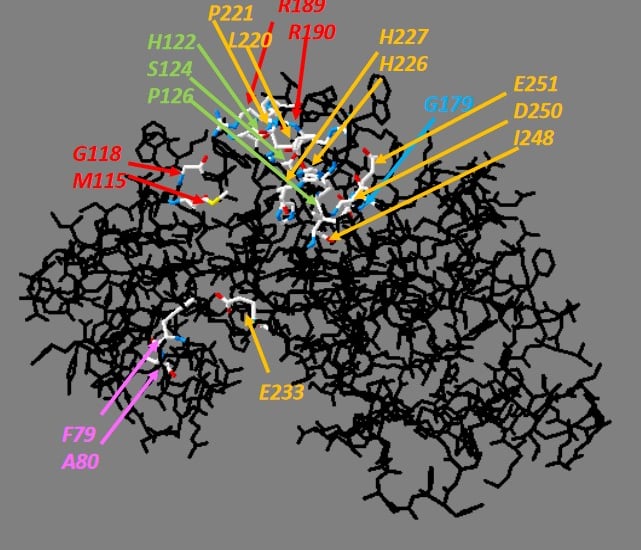
Plants have special, unique Rho-type GTPases (ROPs) and associated signaling components (such as plant-only plant receptor kinases (RLK), RopGEF proteins (PRONE), and ROP effector kinases (RLCKVI_A)). The signaling networks that these proteins form are still poorly known. Until recent years, there has been little data available on the relationship between ROP GTPases and protein phosphorylation, receptor and effector kinases, even though the role of Rho-type GTPases in animal and yeast cells is of fundamental importance in many kinase-mediated signaling pathways. We were the first to demonstrate in our laboratory that ROP GTPases are capable of activating members of a plant-specific kinase family (RLCK VI_A) in vitro, but only in their GTP-bound conformation. In recent years, we have shown that these kinases are involved in similar processes in plants as ROP GTPases, which supports the idea that they can indeed be considered ROP effector kinases. Other experiments have suggested that ROP GTPases themselves may be under phosphorylation regulation and that phosphorylation of the evolutionarily conserved amino acid 74th serine is suitable for inhibiting the activation of the GTPase by RopGEF.
Research
Dedicated to advancing academic knowledge on in vitro plant morphogenesis.
Contact
Follow
feher.attila@brc.hu
+36-62-599-701
© 2025. All rights reserved.